Semiflexible polymers and cytoskeletal filaments
Semiflexible polymers are governed by their bending energy in contrast to flexible polymers, which are governed by entropic tension. Whereas typical synthetic polymers have diameters in the nm-range and are connected by carbon-carbon backbone, which is very flexible, many biopolymers - such as DNA, cytoskeletal filaments (F-actin and microtubules), protein fibers - are relatively large and thick molecules which leads to considerable bending rigidity. The bending rigidity of a polymer can be characterized by its persistence length, which is the ratio of the bending rigidity κ of the polymer and the thermal energy: Lp=κ/kBT. It can be visualized as the typical length scale over which a thermally fluctuating semiflexible polymer changes its orientation; fragments smaller than the persistence length essentially behave as rigid rods. Typical persistence lengths are 50nm for DNA (mechanical contribution, electrostatics increases the stiffness), 10μm for F-actin, or several mm for microtubules.
The reason for a large bending rigidity often is (according to isotropic elasticity theory) the "thickness" of a polymer. Therefore, semiflexible polymers are typically "thick", which makes them suitable for single polymer manipulation and observation. The theoretical description of such single polymer experiments is an important research area.
Another fascinating aspect of the cytoskeletal filaments F-actin and microtubules is their dynamic polymerization behavior which is fueled by the hydrolysis of ATP or GTP. This chemically driven polymerization dynamics provides the basis for cellular force generation and motion and allows for a constant remodelling and, thus, shape changes of the cytoskeleton. This property makes the difference between "dead" synthetic polymers and polymers, which provide the basis for active motion - a basic feature of living matter.
Our research spans from the polymer physics of single semiflexible polymers to structures containing many interacting polymers and from polymers in equilibrium to "living" cytoskeletal polymers with a chemically driven polymerization dynamics:
Single Semiflexible Polymers
The bending rigidity of semiflexible polymers gives rise to some interesting new polymer physics because thermal fluctuations, external forces, or fluctuations in a random potential are also competing with the bending energy. In "conventional" flexible synthetic polymers there would only be competition with the entropic elasticity of a flexible chain.
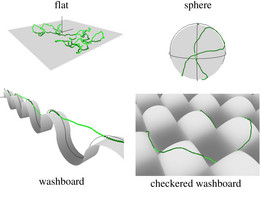
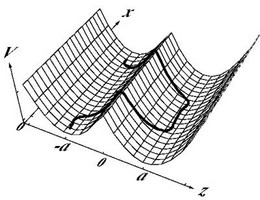
This has consequences, for example, if semiflexible polymers adsorb on structured curved substrates, where adsorption then also involves bending energy.
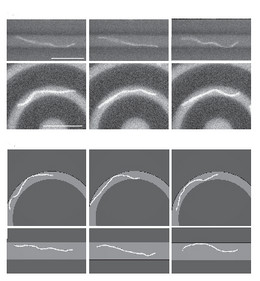
Because many semiflexible polymers are quite "large and thick" (as compared to a typical synthetic polymer), such as cytoskeletal filaments (F-actin, microtubules) or DNA, they are often well suited to be observed and manipulated on a single molecule level. In order to interpret such experiments quantitatively, detailed theoretical models are helpful. Examples are the analysis of thermal fluctuations of single polymers in straight or curved microchannels, the forced desorption from an adhesive substrate, or the dynamic behavior if a polymers is driven over a surface with potential or topographic barriers.
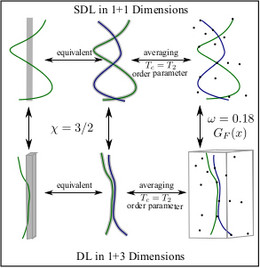
Finally, the fluctuation behavior of semiflexible polymers can provide interesting statistical physics connecting very different problems. For directed lines there exists a mapping from the problem of a single line in a random potential onto the so-called Kardar-Parisi-Zhang (KPZ) equation, which is a very important non-linear equation describing surface growth and whose dynamic critical properties are still not completely clear. We have studied this mapping for stiff lines, which gives some new insight into the problem.
References
- Persistence length
- Persistence length of semiflexible polymers and bending rigidity renormalization
P. Gutjahr, R. Lipowsky, and J. Kierfeld, Europhys. Lett. 76, 994-1000 (2006).
- Persistence length of semiflexible polymers and bending rigidity renormalization
- Adsorption
- Adsorption of finite semiflexible polymers and their loop and tail distributions
T.A. Kampmann and J. Kierfeld, J. Chem. Phys. 147, 014901 (2017). - Controlling adsorption of semiflexible polymers on planar and curved substrates
T. Kampmann, H.-H. Boltz, and J. Kierfeld, J. Chem. Phys. 139, 034903 (2013). - Semiflexible polymer rings on topographically and chemically structured surfaces
P. Gutjahr, R. Lipowsky, J. Kierfeld, Soft Matter 6, 5461-5475 (2010). - Duality mapping and unbinding transitions of semiflexible and directed polymers
J. Kierfeld and R. Lipowsky, J. Phys. A: Math. Gen. 38, L155-L161 (2005). - Unbundling and desorption of semiflexible polymers
J. Kierfeld and R. Lipowsky, Europhys. Lett. 62, 285-291 (2003).
- Adsorption of finite semiflexible polymers and their loop and tail distributions
- Single polymer manipulation
- Characterization of single semiflexible filaments under geometric constraints
S. Köster, J. Kierfeld, and T. Pfohl, Eur. Phys. J. E 25, 439-449 (2008). - Stretching of buckled filaments by thermal fluctuations
K. Baczynski, R. Lipowsky, and J. Kierfeld, Phys. Rev. E 76, 061914 (2007). - Fluctuations of Single Confined Actin Filaments
S. Köster, H. Stark, T. Pfohl, and J. Kierfeld, Biophys. Rev. Lett. 2, 155-166 (2007). - Force-Induced Desorption and Unzipping of Semiflexible Polymers
J. Kierfeld, Phys. Rev. Lett. 97, 058302 (2006). - Point force manipulation and activated dynamics of polymers adsorbed on structured substrates
P. Kraikivski, R. Lipowsky, and J. Kierfeld, Europhys. Lett. 71, 138-144 (2005). - Activated dynamics of semiflexible polymers on structured substrates
P. Kraikivski, R. Lipowsky, and J. Kierfeld, Eur. Phys. J. E 16, 319-340 (2005). - Stretching of semiflexible polymers with elastic bonds
J. Kierfeld, O. Niamploy, V. Sa-yakanit, and R. Lipowsky, Eur. Phys. J. E 14, 17-34 (2004). - Barrier crossing of semiflexible polymers
P. Kraikivski, R. Lipowsky, and J. Kierfeld, Europhys. Lett. 66, 763-769 (2004).
- Characterization of single semiflexible filaments under geometric constraints
- Semiflexible polymers in random media:
- Stiff directed lines in random media
H.-H. Boltz and J. Kierfeld, Phys. Rev. E 88, 012103 (2013). - Localization transition of stiff directed lines in random media
H.-H. Boltz and J. Kierfeld, Phys. Rev. E 86, 060102(R) (2012).
- Stiff directed lines in random media
Bundles and networks of filaments
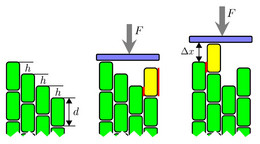
In the cytoskeleton of a cell F-actin filaments typically form bundles or network structures which are hold together by crosslinkers with two "sticky" ends. Here, we study the question, how semiflexible polymers bundle or unbundle from a statistical physics view point. We found that the bundling transition is discontinuous and, thus, rather sharp for stiff polymers.
Moreover, the adhesive energy that is gained in forming a bundle can also be used to exert pushing or zipping forces onto obstacles.
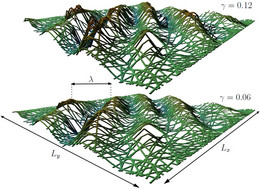
We also investigated networks of semiflexible polymers with respect to wrinkle formation.
References
- Wrinkling of Random and Regular Semiflexible Polymer Networks
P. Müller and J. Kierfeld, Phys. Rev. Lett. 112, 094303 (2014). - Discontinuous Unbinding Transitions of Filament Bundles
J. Kierfeld, T. Kühne, and R. Lipowsky, Phys. Rev. Lett. 95, 038102 (2005). - Zipping mechanism for force generation by growing filament bundles
T. Kühne, R. Lipowsky, and J. Kierfeld, EPL 86, 68002 (2009).
Polymerization kinetics of actin and microtrubules
Cytoskeletal filaments are constantly polymerizing and depolymerizing within the cell. The polymerization dynamics is also chemically driven because ATP (F-actin) or GTP (microtubules) is hydrolyzed within a filament.
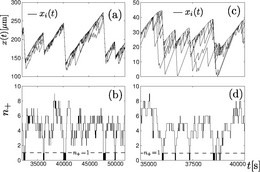
An additional aspect in microtubule polymerization dynamics is their dynamical instability, i.e., the occurence of catastrophes, where the microtubule switches stochastically from a growing state into a state of rapid depolymerization, and rescue events, where it switches back to growth. Catastrophes are associated with a loss of the GTP-cap, which stabilizes the microtubule structure mechanically. We develop models to understand the origin of catastrophe chemomechanically on the dimer level. The dynamic instability has interesting consequences for their ability to generate pushing forces, which becomes limited by catastrophes. It also leads to characteristic collective catastrophes or rescue events if many microtubules cooperate in pushing an obstacle. A cooperative "tug-of-war" of polmerizing and depolyreizing microtrubule ensembles also causes stochastic oscillations in the mitotic spindle.
The polymerization of actin filaments is coupled to their hydrolysis, which determines the ATP-cap strucutre, and to regulating proteins such as profilin, which determines growth speeds. Regulating proteins such as stathmin also play an important role in microtubule dynamics and length regulation.
References
- Chemomechanical Simulation of Microtubule Dynamics With Explicit Lateral Bond Dynamics
M. Schmidt and J. Kierfeld, Front. Phys. 9:673875 (2021). - Bistability and oscillations in cooperative microtubule and kinetochore dynamics in the mitotic spindle
F. Schwietert and J. Kierfeld, New J. Phys. 22 , 053008 (2020). - Feedback Mechanism for Microtubule Length Regulation by Stathmin Gradients
M. Zeitz and J. Kierfeld, Biophys. J. 107, 2860-2871 (2014). - Effects of microtubule mechanics on hydrolysis and catastrophes
N. Müller and J. Kierfeld, Phys. Biol 11, 046001 (2014). - Cooperative dynamics of microtubule ensembles: Polymerization forces and rescue-induced oscillations
B. Zelinski and J. Kierfeld, Phys. Rev. E 87, 012703 (2013). - Dynamics and length distribution of microtubules under force and confinement
B. Zelinski, N. Müller, and J. Kierfeld, Phys. Rev. E 86, 041918 (2012). - Stall force of polymerizing microtubules and filament bundles
J. Krawczyk and J. Kierfeld, EPL 93, 28006 (2011). - Coupling of actin hydrolysis and polymerization: Reduced description with two nucleotide states
X. Li, R. Lipowsky, and J. Kierfeld, EPL 89, 38010 (2010). - Actin Polymerization and Depolymerization Coupled to Cooperative Hydrolysis
X. Li, J. Kierfeld, and R. Lipowsky, Phys. Rev. Lett. 103, 048102 (2009).
Interaction with molecular motors (gliding assays)
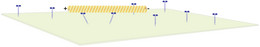
Cytoskeletal filaments also serve as tracks for molecular motor proteins. In gliding assays the motor proteins are immobilized on a surface and pull filaments over the surface. Such gliding assays can be used to study the stepping behavior of single motors, the competittion between different motor types (for exmaple slow and fast motors) transporting a filament, or collective effects from interactions between filaments.
References
- Bifurcation of Velocity Distributions in Cooperative Transport of Filaments by Fast and Slow Motors
X. Li, R. Lipowsky, and J. Kierfeld, Biophys. J. 104, 666-667 (2013). - Critical Motor Number for Fractional Steps of Cytoskeletal Filaments in Gliding Assays
X. Li, R. Lipowsky, and J. Kierfeld, PLoS ONE 7, e43219 (2012). - Active dynamics of filaments in motility assays
J. Kierfeld, K. Frentzel, P. Kraikivski, and R. Lipowsky, Eur. Phys. J. Special Topics 157, 123-133 (2008). - Filament Ordering and Clustering by Molecular Motors in Motility Assays
J. Kierfeld, P. Kraikivski, and R. Lipowsky, Biophys. Rev. Lett. 1, 363-374 (2006). - Enhanced Ordering of Interacting Filaments by Molecular Motors
P. Kraikivski, R. Lipowsky, and J. Kierfeld, Phys. Rev. Lett. 96, 258103 (2006).
Reviews
- Cooperative Behaviour of Semiflexible Polymers and Filaments
J. Kierfeld, P. Kraikivski, T. Kühne, and R. Lipowsky, in Traffic and Granular Flow '05 , 239-249, Springer (2006). - Modelling semiflexible polymers: shape analysis, buckling instabilities, and force generation
J. Kierfeld, K. Baczynski, P. Gutjahr, T. Kühne and R. Lipowsky, Soft Matter 6, 5764-5769 (2010).